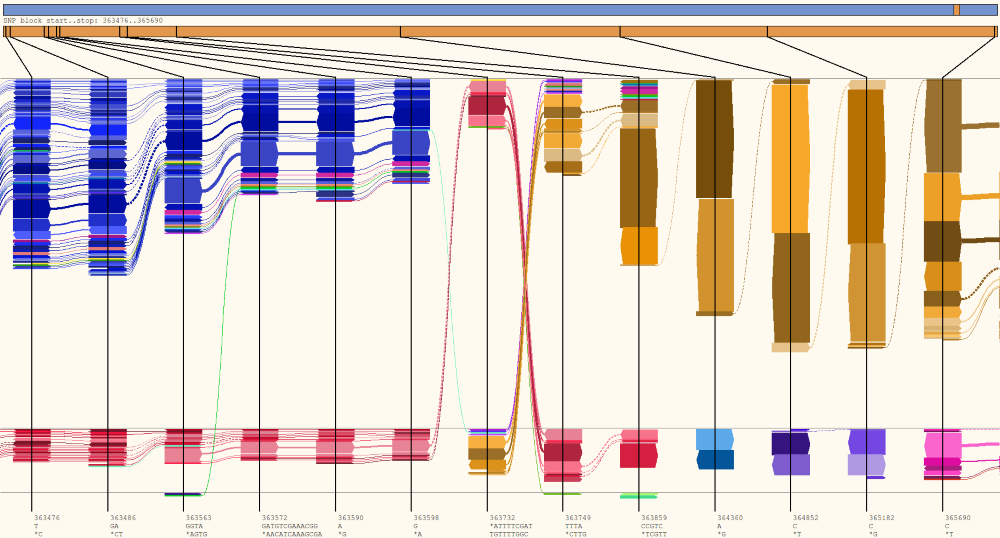
HapFlow
HapFlow is a python application for visualising haplotypes present in sequencing data. It identifies variant profiles present and reads and creates an abstract visual representation of these profiles to make haplotypes easier to identify.
HapFlow was developed in collaboration with Dr. Adam Polkinghorne and Dr. Nathan Bachmann at the University of the Sunshine Coast .
For installation instructions and binaries go to Downloads.
For a comprehensive guide to using HapFlow please visit the wiki.
For a quick start guide and more in depth tutorials go to Tutorials.
For information on how to cite HapFlow visit Citing.
If you have any problems with HapFlow or and feature suggestions post here.
I will endeavour to get back to you as soon as possible.
Email: mjsull@gmail.com
Follow @mjsull1
Updates
-
Release version.
-
Website update.
-
Version 1.1.0 release.
Included option to define OTUs and write reads conforming to an OTU to BAM files.
-
Updated website.